Up to half of the population in the United States (opens in new tab) and United Kingdom (opens in new tab) will be diagnosed with cancer at some point in their lives. Of those, half will be treated with radiotherapy (RT), often in combination with other treatments such as surgery, chemotherapy, and increasingly immunotherapy. Radiotherapy involves focusing high-intensity radiation beams to damage the DNA of deep-seated cancerous tumors while avoiding surrounding healthy organs (known as organs at risk or OARs). Around 40% of successfully treated cancer patients undergo some form of radiotherapy, which shows how critical this tool is to cancer treatment regimens.
Project InnerEye is research conducted in the Health Intelligence team at Microsoft Research Cambridge (UK) that is exploring ways in which machine learning (ML) has the potential to assist clinicians in planning their radiotherapy treatments so that they can spend more time with their patients. In September 2020, we released the InnerEye open-source deep learning toolkit. Our latest peer-reviewed findings, published in the JAMA Network Open article titled “Evaluation of Deep Learning to Augment Image Guided Radiotherapy for Head and Neck and Prostate Cancers,” address the question:
Can machine learning (ML) models achieve clinically acceptable image segmentation in radiotherapy planning and reduce overall contouring time?
Planning radiotherapy treatment can be a lengthy process. It starts with a 3D CT (Computed Tomography) imaging scan of the part of the body to be targeted. These CT images come in the form of stacks of 2D images, dozens of images deep, each of which must be examined and marked up by a radiation oncologist or specialist technician. This process is called contouring. In each image, an expert must manually draw a contour line around the tumors and OARs in the target area using dedicated computer software.
For complex cases, this can take several hours in the planning of a single patient’s treatment. This image segmentation tasks consumes significant time and resources in the cancer treatment pathway for radiotherapy, which increases the burden on clinicians and the final cost to hospitals. As this task is subjective, there can be significant variability across experts and institutions where protocols and patient demographics vary. This is a limitation to the use of imaging in clinical trials and can introduce variability in patient care.
Our work shows that clinicians using ML assistance can segment images up to 13 times faster than doing it manually, with an accuracy that is within the bounds of human expert variability.
Sourcing information from 8 clinical centers to build robust and generalizable machine learning models
One of the barriers to the uptake of ML in clinical use across different hospitals is that most models are only trained on a dataset from a single institution and focus on a single task. This lack of generalizability can reduce the potential utility of ML in the real world, due to the model robustness across different institutions. To overcome this, we have developed generic models, trained on anonymized data from eight different clinical centers across Australia, Europe, New Zealand, North America, and South America. Models were trained on a dataset of 519 pelvic 3D CT planning scans and 242 of the head and neck, acquired as part of the treatment dose planning process. All identifying information was removed from the data by the clinical sites prior to the transfer to Microsoft Research1.
The de-identified images were then manually annotated by two clinically trained expert readers and two radiation oncologists. Two datasets were used for different purposes in this research: a main dataset and an external dataset. In the main dataset, the ML segmentation models were trained on data from five clinical sites to automatically delineate 15 different target structures. The external dataset comprised data not used for training from three of the clinical sites, which allowed for blind testing of the ML models.
1 A defined list of tags was used indicating which are retained, hashed, or randomized in the case of the dates and times. Any tags that were not in the list were removed. Tags were only retained where related to image geometry and scanner settings and other non-identifiable tags (such as gender) that were necessary to process the data.
Working toward the end goal of saving clinicians time and resources
We performed an evaluation of the potential clinical utility of ML in the radiotherapy planning pathway by comparing the time taken for the end-to-end image segmentation task performed manually by clinicians, with the time taken when clinicians use the ML model to assist them in marking up images. This provides key insights into how ML might be applied to reduce clinician workload, overall planning time, and hospital costs.
The image segmentation model is a state-of-the-art convolutional neural network based on a 3D U-Net architecture, with approximately 39 million trainable parameters.
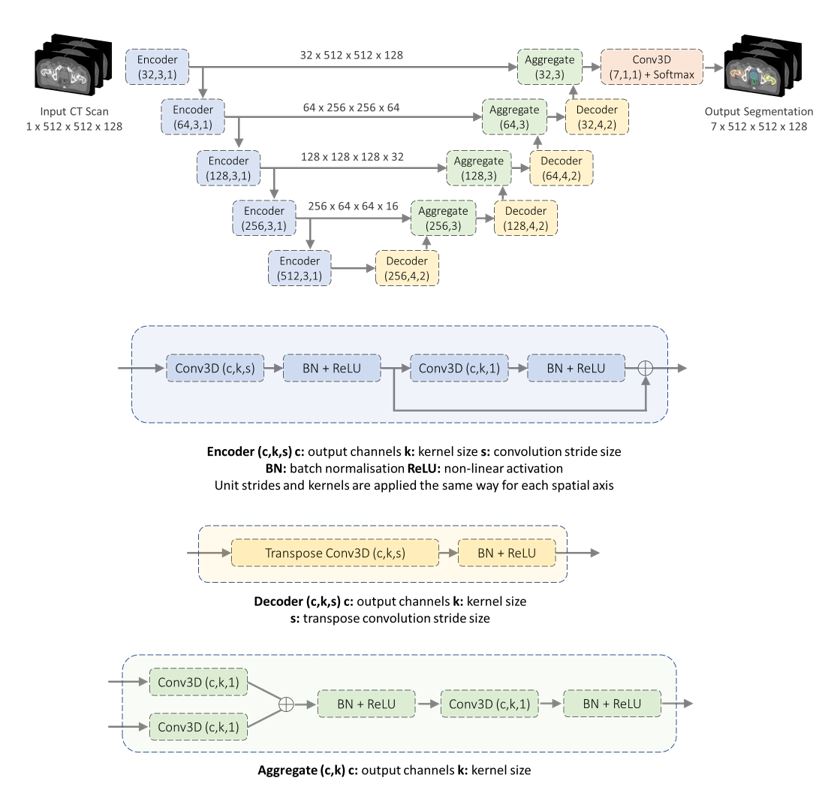
We used Microsoft Azure Machine Learning to allow us to easily develop and train our models across 20 NVIDIA Tesla V100 GPUs. See our paper for how we used mixed-precision representations and pipeline parallelism to reduce memory requirements, lower time to solution, and improve inference speed of the model.
Is the ML model usable in radiotherapy practice?
One way of thinking about this is to check how the model compares with the difference of interpretation between expert clinicians performing the same task. To test this, we performed an inter-observer variability (IOV) study using 10 test images each for pelvis and head-and-neck cases, comparing the variability between three expert clinicians.
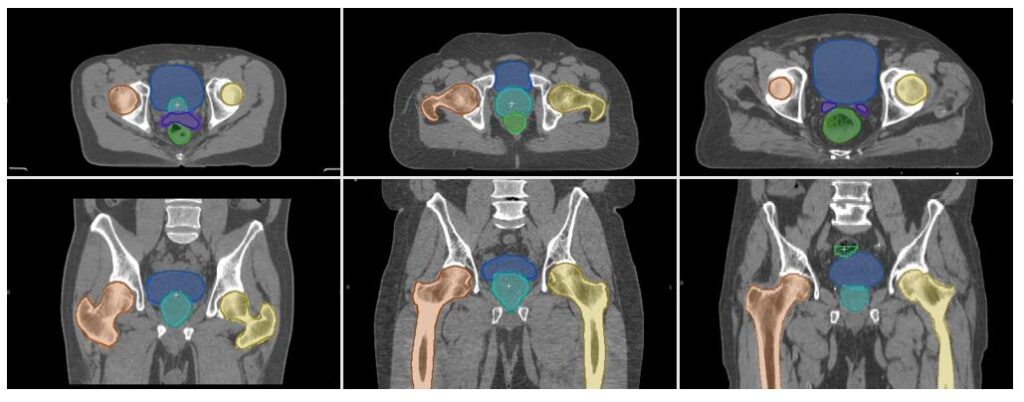
To create ground truth segmentations, we collected manually generated contours from multiple experts and aggregated them using a majority voting rule. For instance, in the case of three experts, if two of the experts indicated that a particular voxel belonged to the prostate, then a prostate label was assigned regardless of the third labeler’s opinion. For the IOV study we used a volumetric measure called the Dice score (opens in new tab) that compared the overlap between segmented structures in pairs of images. Perfectly overlapping structures have a Dice score of 100%, while a Dice score of 0% corresponds to no overlap. We also measured the distance between the surface contour of the manual and automatic segmentations as additional metrics: Hausdorff distance and mean surface-to-surface distance.
Spotlight: Event Series
We used two widely used statistical measurements, Cohen’s Kappa and Fleiss’ Kappa for single and multiple annotators respectively, to assess the agreement between contours generated by the model and expert readers. After plotting these results visually, we learned that similarity scores when compared with ground truth are on par with expert IOV in contouring, giving us some confidence that errors in the ML model fall within the variability of the three experts involved in the IOV study. See our paper for the statistical agreement results, such as Bland-Altman plots.
We also wanted to see if the ML model might be usable on datasets not used in training and applicable for more widespread use. So we tested it on external datasets from three clinical sites, separate from those used for training, each of which had different CT imaging protocols, scanner hardware, and patient groups. This would give us confidence in the generalizability of the model beyond the sites supplying training data. We used the Mann-Whitney U test to measure the model performance difference across datasets. Our observations of the segmentation errors tended to occur in the superior and inferior extent of tubular structures and in the interface between adjacent organs. However, we have not observed any inconsistencies that, if not corrected, could lead to significant errors in a treatment plan, as evidenced by the surface distance results. This is because the proposed postprocessing method does not allow inconsistencies at a distance from the anatomical structure by design.
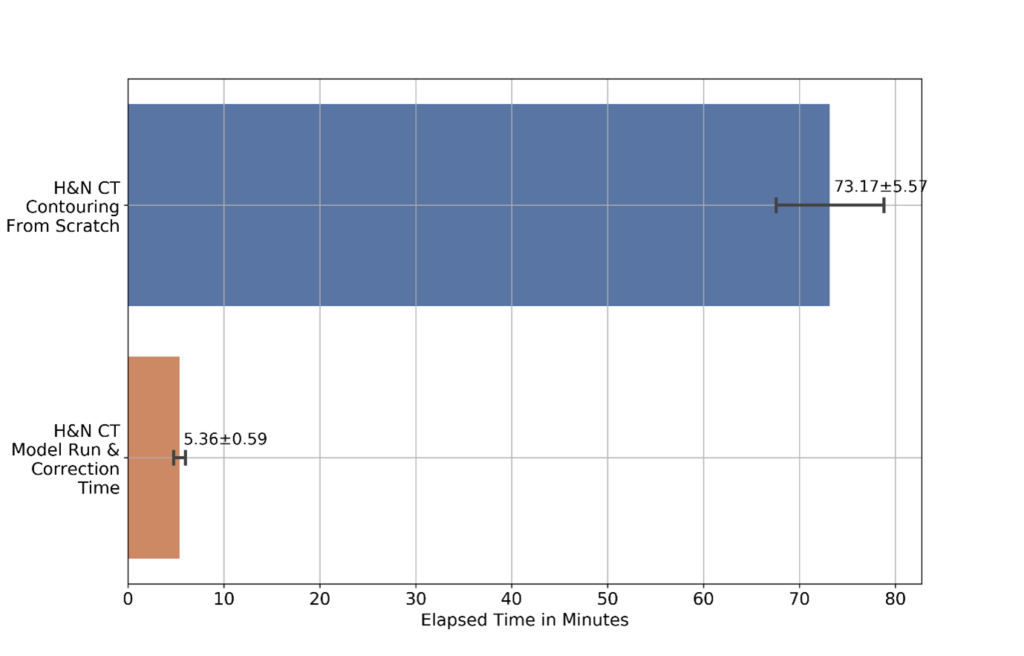
Our results show that the ML model greatly reduces the time it takes for end-to-end image segmentation and annotation in radiotherapy. This included both the time to draw segmentation contours on the image and time to correct inaccuracies in the automated (or semi-automated) system. Figure 4 shows the time taken for a radiation oncologist to perform image contouring manually, compared with using the ML model, including the time for the expert to inspect and update the contours to ensure clinical accuracy. The time taken for the ML model to perform inference was only 23 ± 3 seconds in a full input CT scan. For the radiation oncologists in this specific research study, the time taken is shown to be reduced by over 90% for head and neck image segmentation.
Creating an end-to-end deployment framework for use in clinics
Our reproducible ML model and work creates an opportunity for easy and widespread adoption of auto-segmentation models into existing radiotherapy workflows. However, creating an ML model that performs well enough to be clinically useful does not necessarily mean that it can be deployed successfully in the clinic. The additional engineering and infrastructure required to integrate it into a clinical setting is significant. To help bridge this gap between research and application deployment, the InnerEye team has been working with Microsoft Azure over the last three years to create an end-to-end framework for both edge and cloud deployment using the industry-standard Digital Imaging and Communications in Medicine (DICOM) image format.
In the proposed workflow shown in Figure 4, CT scans are acquired from patients as they attend preparations for radiotherapy treatment. These scans are initially stored at the hospital’s image database and later securely transferred via the gateway to the auto-segmentation platform in the cloud after anonymizing them. Once the segmentation process is completed, resultant files are uploaded back to the hospital’s image database, creating a seamless clinical workflow where clinicians can review and refine contours in their existing contouring and planning tools.
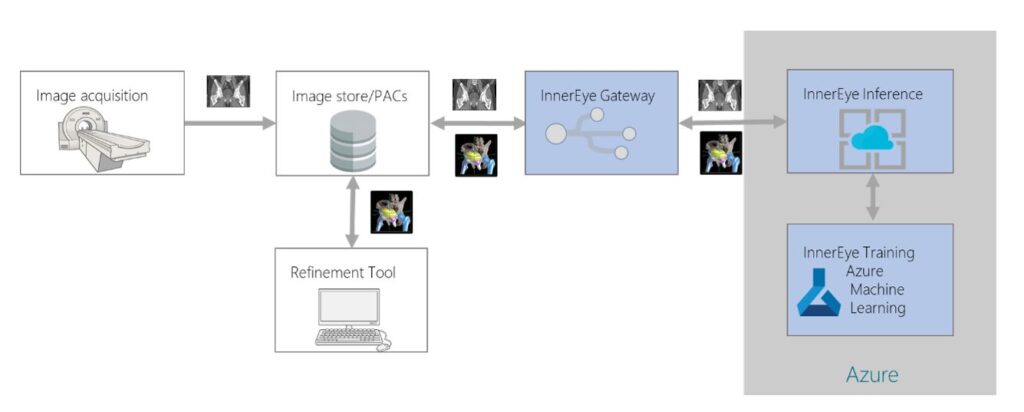
Clinical utility of ML for radiotherapy
This most recent work demonstrates the potential for our InnerEye research in the clinical world. We have shown how we:
• trained ML models that can be easily integrated into current radiotherapy practices (with approval from the appropriate regulatory agencies) and have accuracy within the bounds of human expert variability;
• tested the robustness of the model when applied to images from clinical sites with different protocols and imaging hardware;
• indicated potential time savings for complex radiotherapy planning clinical workflows of over 90%; and
• developed underlying cloud platform technology that could be used for seamless integration into existing clinical workflows.
While the ML models have been shown to perform well enough to be relevant for clinical practice, it is imperative that clinicians and experts remain in the loop to assess accuracy and clinical significance. The ability for experts to manually correct the model outputs is a necessary component of the ML-augmented radiotherapy workflow.
We hope our latest work contributes to addressing the practical challenges of scalable adoption of ML across healthcare systems and opens possibilities for new radiotherapy treatments to become mainstream. By making the source code used in this study publicly available as open-source software in the InnerEye Deep Learning Toolkit on GitHub (opens in new tab), we are making our research more reproducible and empowering researchers and organizations to build on this work by training and deploying their own ML models, using their own datasets.
-
VIDEO
Tech Minutes: Inner Eye
Javier Alverez explains how we’re using state of the art machine learning technology to build innovative tools for the automatic, quantitative analysis of three-dimensional medical images.
We are excited to see how our work will be built upon to improve the experience of clinicians planning radiotherapy and enhance cancer treatment for patients at centers around the world.